Note
Go to the end to download the full example code.
Interactive tools for image visualization
Learn about 3D and 4D image visualization tools from PyEPRI (require PyEPRI >= 1.1.1).
Interactive 3D image viewers
Let’s start by generating some data by rerunning the simplified 3D image reconstruction example on the fusillo sample dataset.
# --------------------- #
# Import needed modules #
# --------------------- #
import matplotlib.pyplot as plt # tools for data visualization
import pyepri.apodization as apodization # tools for creating apodization profiles
import pyepri.backends as backends # to instanciate PyEPRI backends
import pyepri.datasets as datasets # to retrieve the path (on your own machine) of the demo dataset
import pyepri.displayers as displayers # tools for displaying images (with update along the computation)
import pyepri.monosrc as monosrc # tools related to standard EPR operators (projection, backprojection, ...)
import pyepri.multisrc as multisrc # tools related to multisources EPR operators (projection, backprojection, ...)
import pyepri.spectralspatial as ss # tools related to spectral spatial EPR operators (projection, backprojection, ...)
import pyepri.processing as processing # tools for EPR image reconstruction
import pyepri.io as io # tools for loading EPR datasets (in BES3T or Python .PKL format)
# ---------------------------- #
# Create a numpy (CPU) backend #
# ---------------------------- #
backend = backends.create_numpy_backend()
# ----------------------------------------------------------------- #
# Load one demonstration dataset (fusillo pasta soaked with TEMPOL) #
# ----------------------------------------------------------------- #
dtype = 'float32' # use 'float32' for single (32 bit) precision and 'float64' for double (64 bit) precision
path_proj = datasets.get_path('fusillo-20091002-proj.pkl') # or use your own dataset, e.g., path_proj = '~/my_projections.DSC'
path_h = datasets.get_path('fusillo-20091002-h.pkl') # or use your own dataset, e.g., path_h = '~/my_spectrum.DSC'
dataset_proj = io.load(path_proj, backend=backend, dtype=dtype) # load the dataset containing the projections
dataset_h = io.load(path_h, backend=backend, dtype=dtype) # load the dataset containing the reference spectrum
B = dataset_proj['B'] # get B nodes from the loaded dataset
proj = dataset_proj['DAT'] # get projections data from the loaded dataset
fgrad = dataset_proj['FGRAD'] # get field gradient data from the loaded dataset
h = dataset_h['DAT'] # get reference spectrum data from the loaded dataset
# ----------------------------------------------------- #
# Configure and run TV-regularized image reconstruction #
# ----------------------------------------------------- #
delta = .1; # sampling step in the same length unit as the provided field gradient coordinates (here cm)
out_shape = (50, 25, 25) # output image shape (number of pixels along each axis)
lbda = 500. # regularity parameter (arbitrary unit)
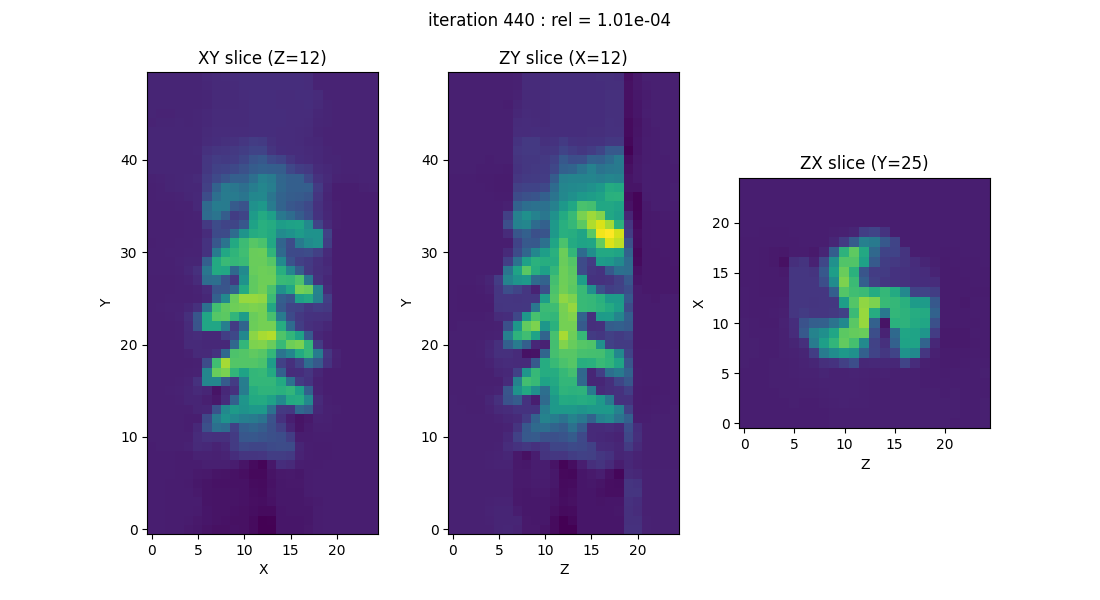
displayer = displayers.create_3d_displayer(nsrc=1, figsize=(11., 6.), display_labels=True)
out = processing.tv_monosrc(proj, B, fgrad, delta, h, lbda, out_shape, backend=backend,
init=None, tol=1e-4, nitermax=500, eval_energy=False,
verbose=False, video=True, Ndisplay=20, displayer=displayer)
plt.show() # to keep the display persistent when the code is executed as a script
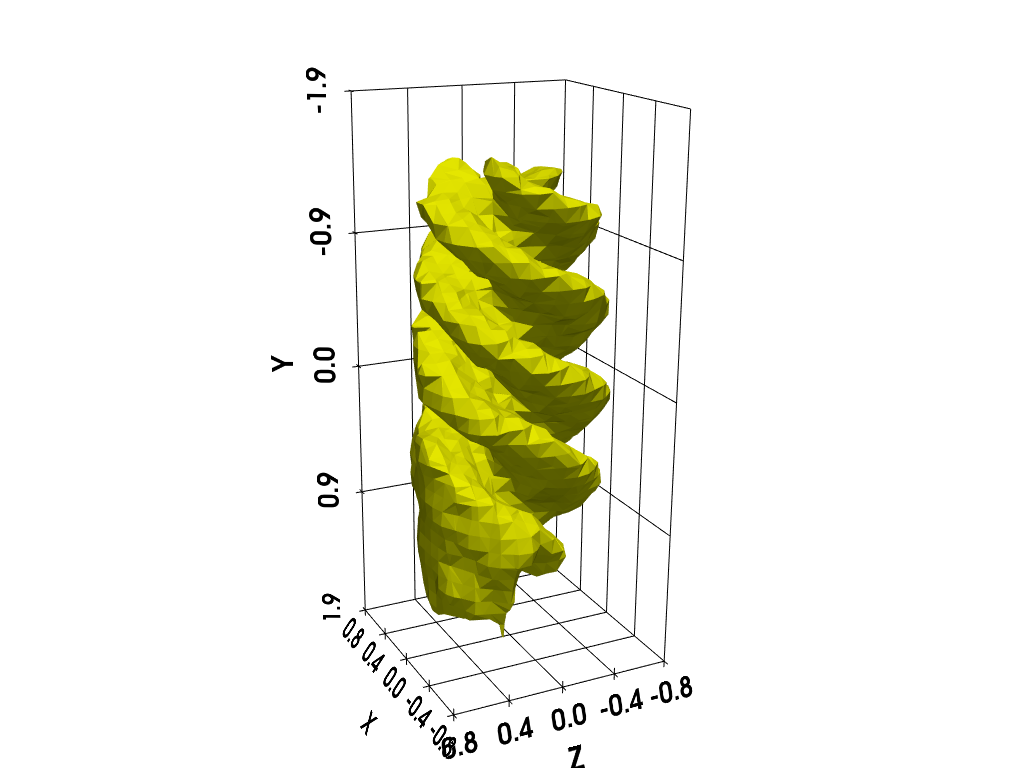
Basic Isosurface rendering (PyVista)
You can use the following code to “manually” compute and display an isosurface of the 3D image using PyVista.
# additional imports
import numpy as np
import pyvista as pv # tools for rendering 3D volumes
from pyepri.utils import otsu_threshold # used for automatic isovalue computation
# prepare isosurface display
xgrid = (-(out_shape[1]//2) + np.arange(out_shape[1])) * delta # X-axis (horizontal) sampling grid
ygrid = (-(out_shape[0]//2) + np.arange(out_shape[0])) * delta # Y-axis (vertical) sampling grid
zgrid = (-(out_shape[2]//2) + np.arange(out_shape[2])) * delta # Z-axis (depth) sampling grid
x, y, z = np.meshgrid(xgrid, ygrid, zgrid, indexing='xy')
grid = pv.StructuredGrid(x, y, z)
# compute isosurface
vol = np.moveaxis(backend.to_numpy(out), (0,1,2), (2,1,0))
grid["vol"] = vol.flatten()
isolevel = otsu_threshold(vol) # change the value here if needed (e.g., isolevel = .5 * vol.max())
contours = grid.contour(isolevel)
# display isosurface
cpos = [(-8.2, -3.1, 4.1), (0.0, 0.0, 0.0), (0.3, -0.95, -0.14)]
p = pv.Plotter()
p.camera_position = cpos
labels = dict(ztitle='Z', xtitle='X', ytitle='Y')
p.add_mesh(contours, show_scalar_bar=False, color='#f7fe00')
p.show_grid(**labels, bounds=[-0.8, 0.8, -1.9, 1.9, -0.8, 0.8])
p.show()
Interactive slices displayer (Matplotlib)
Use pyepri.displayers.imshow3d() to display the isosurface
and slices of the 3D images using PyVista. Make sure the Matplotlib
window has focus, then press the h key on your keyboard to
display the list of interactive commands in your Python console.
Note: the slice selectors (sliders) won’t be active on this online documentation, but they should work when running the code on your system.
fig = displayers.imshow3d(backend.to_numpy(out), xgrid=xgrid, ygrid=ygrid, zgrid=zgrid, units='cm', figsize=(14.5, 8.8))
plt.show() # to keep the display persistent when the code is executed as a script

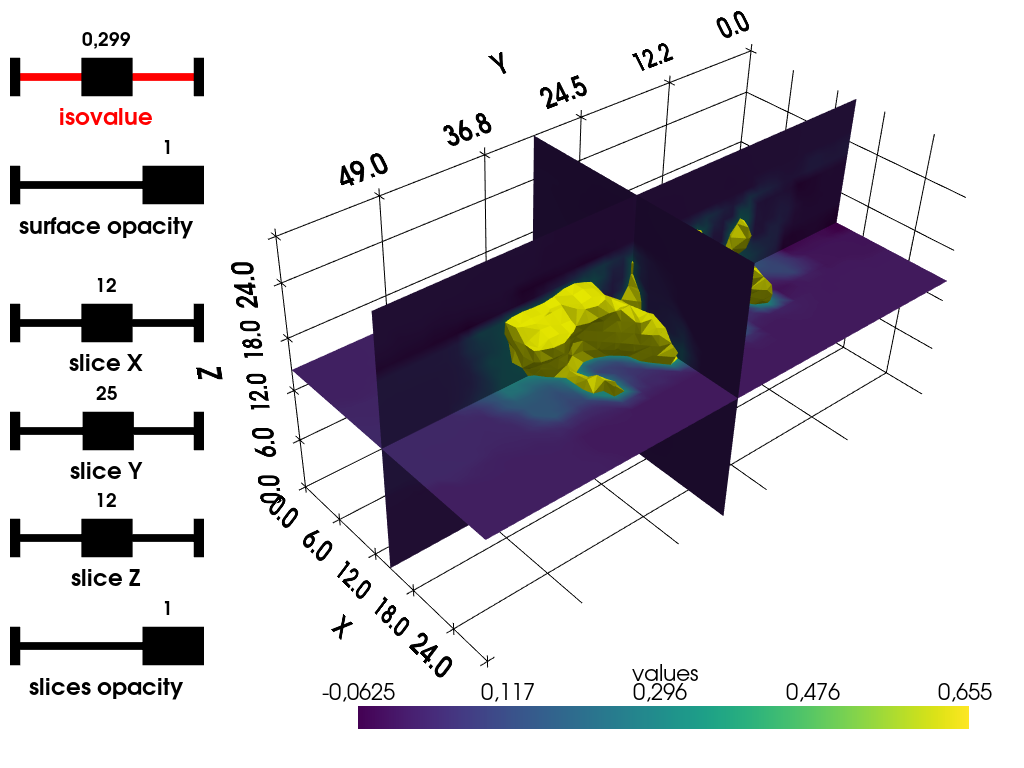
Interactive isosurface + slices displayer (PyVista)
Use pyepri.displayers.isosurf3d() to display isosurface and
slices of the 3D images using PyVista, press key h on your
keyboard to display the list of interactive commands into your
Python console.
Notes:
The slice images are rendered using bilinear interpolation, which results in overly smooth rendering near the edges of the objects.
The widgets (here, sliders) will be disabled on the interactive view of this online documentation, but they should work when running the code on your system (either using Python console or Jupyter Notebook).
plotter = displayers.isosurf3d(backend.to_numpy(out))
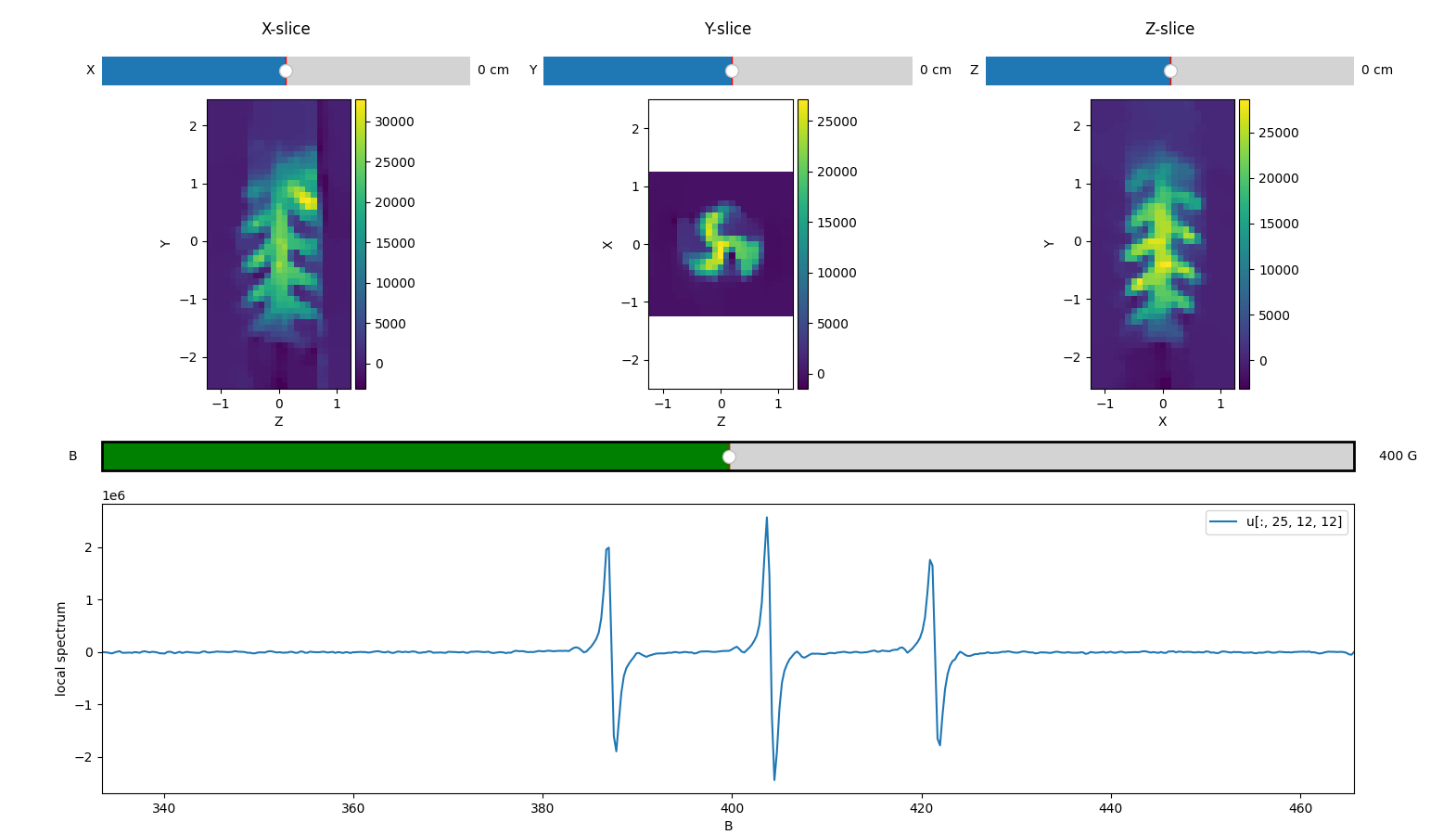
Interactive 4D spectral-spatial images viewer (matplotlib)
Below, we compute a 4D spectral spatial image by simply multiplying each voxel of the 3D image (concentration mapping) computed above by the reference spectrum.
im4d = h.reshape(-1, 1, 1, 1) * out.reshape((1, *out.shape)) # im4d[:, i, j, k] = spectrum
# at voxel location [i, j, k]
You can use pyepri.displayers.imshow4d() to display the
contents of the 4D image. The spectrum corresponding to the voxel
currently under the mouse cursor is shown in the lower panel. When
you click on a voxel, its spectrum is preserved. Press h while
the window is focused to display the full list of interactive
commands.
Note: again, the slice selectors (sliders) won’t be active on this online documentation, but they should work when running the code on your system.
fig = displayers.imshow4d(backend.to_numpy(im4d), xgrid=xgrid, ygrid=ygrid, zgrid=zgrid,
Bgrid=backend.to_numpy(B), B_unit='G', spatial_unit='cm',
figsize=(15.7, 9))
plt.show() # to keep the display persistent when the code is executed as a script
Total running time of the script: (0 minutes 4.711 seconds)
Estimated memory usage: 542 MB